|

from
StarchildUK Website
Scientific Testing - Ancient DNA Test
The Starchild’s mitochondrial DNA was relatively easy to recover and
showed it had a human mother (expected if it is an alien hybrid),
but its nuclear DNA, the part that would reveal its father’s genetic
heritage, couldn’t be recovered with current primers. We were
advised to wait for primers to become more efficient. We were also
advised to investigate its bone chemistry because in conducting the
DNA tests, some intriguing discoveries were made.
1. The bone was significantly
harder to cut that it should have been.
2. There was a stronger-than-usual smell of "burning
bone" when cutting it.
3. When put into EDTA, the normal solvent for human bone,
the Starchild should have dissolved in a week or less since it
is less than half as thick as normal human bone. 10 weeks later
the Starchild bone had not dissolved a bit.
4. When a strong detergent known as ’tween 20’ was added
to the mix, the Starchild bone dissolved completely, overnight,
down to a thin layer of residue. Unexpected.
Thus, its chemistry seemed to be unusual
enough to warrant a full-scale investigation. For most of 2004 we
did precisely that, and now we have some astonishing results that
have scientific merit and investigative significance of the highest
magnitude.
Analysis
The main analysis so far is contained in
3 reports:
PRELIMINARY ANALYSIS OF A
HIGHLY UNUSUAL HUMAN-LIKE SKULL
Dr. Ted J. Robinson
M.D., L.M.C.C., F.R.C.S (c)
The skull in question has a provenance that is not verified at
present. That situation may change in time, but for now all that can
be said with certainty is that the skull is real, it is comprised of
calcium hydroxyapatite (the essence of all mammalian bone), its
parts are configured "naturally" (not cobbled together or in any
other way hoaxed), and it presents numerous physical anomalies that
do not conform to standard skull norms.
The skull remained in my possession in Vancouver, B.C., for the
better part of one year. I was given complete discretion to study it
in any way I saw fit. My analysis derives from extensive examination
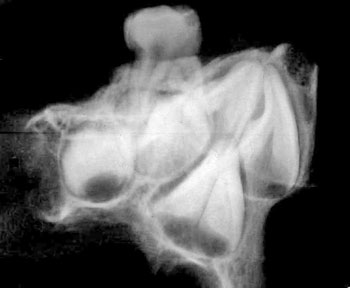
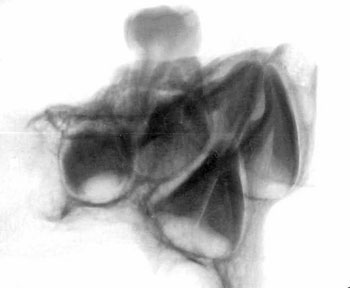
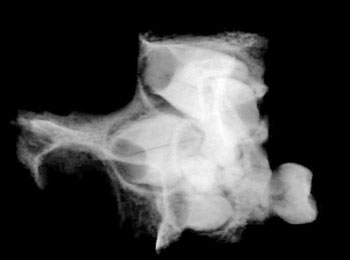
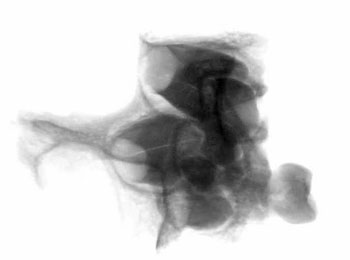
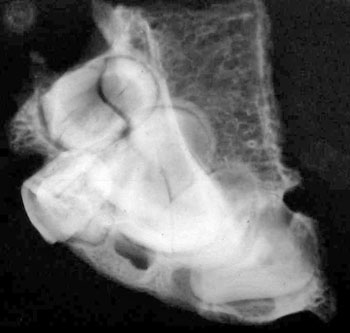
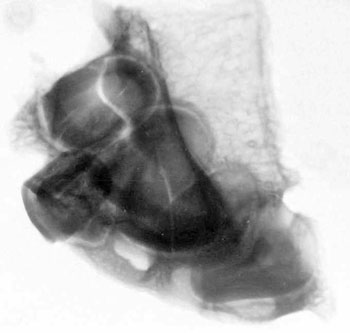
of the skull itself, combined with analysis of X-rays and CAT scans.
I have shared these data with colleagues who have given opinions
that will be mentioned in this document as their input becomes
relevant.
In general, the skull has the basic components of a human skull:
i.e., a frontal bone, two sphenoids, two temporals, two parietals,
and an occipital. However, these bones have been markedly
reconfigured from the "normal" shapes and positions such bones
usually have. In addition, the bone itself has been reconstituted to
an equally marked degree, being somewhat less than half as thick as
normal human bone, with a corresponding weight of roughly half
normal. The reconfigurations and the reconstitution are uniform
throughout all axes and in all planes of the skull. There is no
asymmetrical warping or irregular thinning that is the hallmark of
typical human deformity.
The morphology of this skull is so highly unusual as to be unique in
my forty years of experience as a medical doctor specializing in
plastic and reconstructive surgery of the cranium. Because of its
uniqueness, I undertook an extensive review of current literature on
craniofacial abnormalities, which failed to uncover a single similar
example. In short, it seems to be not only unique in my personal
experience, but also unique throughout the past history of worldwide
study of craniofacial abnormalities. This is significant.
Specialists who examined the skull and associated X-rays and CAT
scans were:
-
Dr. Fred Smith, Head of
Pediatrics, Children’s Hospital, New Orleans, La.
-
Dr. David Hodges, Radiologist,
Royal Columbian Hospital, New Westminster, B.C.
-
Dr. John Bachynsky, Radiologist,
New Westminster, B.C.
-
Dr. Ken Poskitt, Pediatric
Neuroradiologist, Vancouver Children’s Hospital
-
Dr. Ian Jackson, (formerly of
Mayo Clinic), Craniofacial Plastic Surgeon, Michigan
-
Dr. John McNicoll, Craniofacial
Plastic Surgeon, Seattle
-
Dr. Mike Kaburda, Oral Surgeon,
New Westminster, B.C.
-
Dr. Tony Townsend,
Ophthalmologist, Vancouver
-
Dr. Hugh Parsons,
Ophthalmologist, Vancouver
-
Dr David Sweet, Forensic
Odontologist, Vancouver
Dr David Hodges, a radiologist, stated
that the suture lines were open and growing at the time of death.
Dr.David Sweet, an internationally renowned forensic pathologist at
the University of British Columbia, was of the opinion that the
skull was that of a 5-6 year old, based upon the dentition in the
right maxillary fragment[1].
Though some specialists who looked at the skull disagreed, I have
always supported Dr Sweet in his belief that this was the skull of a
5-6 year old child.
Dr. Bachynsky noted that there is no evidence of erosion of the
inner table of the skull. Such erosion would be consistent with a
diagnosis of hydrocephaly, so this condition can safely be ruled out
as a cause of the abnormalities expressed. Hydrocephaly also causes
a widening of the sutures, again not expressed here. There was
consensus agreement to both of these observations by other experts
conversant with these features.
Dr. Kaburda carried out special three-dimensional X-rays which
measure certain fixed points in any skull, allowing for comparison
of any particular skull to the established norm. These accumulated
results were compared to a statistical analysis of 100 human skulls.
This skull was found to be more than ten (10) standard deviations
outside the norm, i.e. the statistical center of a Bell curve. This
is another strong indication that the skull in question is unlike
anything previously seen or investigated.
Doctors Townsend and Parsons examined the orbital cavities and
concluded that the being may well have been sighted, but if so, its
visual structures deviated strongly from the norm. The cavities,
while astonishingly symmetrical, were less than 50% normal depth.
The optic foramen, which carries the optic nerve from the brain
through the orbital bone to the eye, is nearly an inch lower than it
would be in a normal human skull. However, attachment points for the
muscles that control an eyeball’s movements were still to be felt on
the inner surface of the orbit, indicating that a ball rather than
some other mechanism was its most likely expression.
If indeed these sockets held eyeballs, those of normal size would
have greatly protruded from the face, creating a serious liability
of damage during routine activity. Because the eyeballs occupy a
position lower in the face than is normal, and they rest in a socket
markedly reduced in rectilinear shape and depth, they would have
been significantly reduced in size. In either case, however, large
eyeballs or small, they would require upper lids three or four times
more extensive than normal upper lids to be lubricated in the manner
necessary for human eyeballs to function properly.
Doctors Hodges and Poskitt found the brain inside the skull was
abnormally large. This was determined by lining the intracranial
cavity with a plastic bag that was then filled with Niger birdseed.
This gave a size of 1600 cubic centimetres, which is 200 c.c. larger
than the typical adult size of 1400 c.c. This is even more unusual
because the size of the skull compares most favourably with a small
adult or a child of about 12 years old. This extra brain capacity is
apparently due to the deep shallowing of the eye sockets, a total
lack of frontal sinuses (not even vestigial bumps are discernable),
and significant bossing (expansion) of the upper rear of both
parietals.
In any case, they observed, the extreme slant of the rear parietals
and the occipital bone challenges whether this skull could have
contained typical brain matter, and casts further doubt that its
cerebellum was typical. In a normal skull, the cerebellum rests at
the base of the cerebrum, supported by the internal occipital
protuberance and the twin flares of the sagittal sulcus and the
transverse sulcus. With this support mechanism, over the course of a
lifetime the cerebrum’s weight does not press down onto the
cerebellum and distend it such that it will cease to function
properly. In this unique skull, however, the entire weight of the
brain slants directly down on the area that should hold its
cerebellum. Instead of the rounded area typically present for
support, there is a wedge-shaped area of perhaps one-quarter of
normal. Furthermore, the internal protuberance and sulcus ridges are
significantly reduced. What effect would the weight of a notably
amplified brain have on an unsupported cerebellum carried into
adulthood? It presents a genuine conundrum.
Personally, I was most concerned with determining how the rear of
the skull could have become so flattened, from the atypical fossa
(depression) in the sagittal suture between the parietals, down to
the foramen magnum opening. This could not have been caused by any
kind of flattening or binding device because the surface of the
occipital reveals the subtle convolutions inevitably present in
unaltered skulls. Skulls that undergo any kind of shaping technique
will always reveal such technique with a distortion of the bone
surface. Lacking even a hint of evidence of shaping, and of any
unnatural or premature fusing of any sutures, it is entirely safe to
say that the extreme flattening of the skull was caused by its
natural growth pattern and is not artificial. This too is
significant.
Another of my concerns is that the external occipital protuberance (inion)
is absent from its notable position in the center of the occipital
bone, and indeed is represented by an actual slight fossa
(depression) in the surface. (As mentioned earlier, the same is true
for its internal counterpart, which has been greatly reduced.) It
seems clear that the neck of this being attached to its skull much
lower than in a normal skull, centered under the balance point for
both lateral and medial flexion. Even more unusual, the neck itself
seems to have a circumference somewhere in the range of 50% of usual
neck volume, which presents yet another example of the thorough
uniqueness of this specimen.
In addition to lacking frontal sinuses, there is no sign of the brow
ridges evident in normal skulls. Its upper orbits are thin edged
rather than rounded. Its zygomatic arches are greatly reduced and
significantly lowered from their usual positions. Its mastoid
processes are less than normal, as are all connective points for the
lower face (which would attach to the coronoid process and condylar
process of the missing mandible). Based on these observations, its
lower face may have been as much as 50% reduced from normal. On the
other hand, its inner ears are noticeably larger than normal, again
pushing into the range of 50% larger. This is also true for the
condyles abutting the spinal atlas.
A detached upper right maxilla contains two molars [recent note: one
has been lost to testing]. Tooth wear on the molars indicates
maturity was reached, yet another set of teeth are present in the
maxilla and appear ready to take the place of those mature teeth
when and if they are lost or are no longer useful. The question of
age at death remains open.
Carbon 14 Dating has shown the Human Skull to be 900 years old ± 40
years[2].
These and other mysteries about this skull await further analysis by
other experts wishing to help determine its origin and history.
References:
[1] Dr Matthew Brown, a
Dentist in London, made close-up x-rays images of the maxilla in
September 2004. He states that the roots of unerupted teeth are
consistent with those of a child who was about 4˝ yrs old.
[2] Carbon 14 dating was also carried out on Starchild
Skull Bone in July/August 2004 which produced the same result -
900 years old ± 40 years
Go Back
Report on Maxilla and Teeth
X-Rays
Dr Matthew Brown, DDS.
In September 2004, some New X-rays of the Starchild Teeth were
completed. The report and results are shown below.
|
27 October 2004
Examination of infantile right maxilla, anterior fragment, with
regard to dentition.
Visual:
One tooth present (upper right first deciduous molar (54*)). Alveoli
(sockets) of upper right first deciduous incisor, second deciduous
incisor, deciduous canine and deciduous second molar present (51,
52, 53, 55). Anterior/mesial wall of upper right permanent first
molar (16) crypt visible. The crown of 54 shows occlusal wear facets
and enamel lamellae.
Radiographic.
Crowns of upper right permanent
. central incisor (11)
. lateral incisor (12)
. canine (13)
. first premolar (14)
. second premolar (15)
visible, with initial root development on the central incisor;
crowns of the remaining teeth at an incomplete stage of development.
Interpretation:
The absent teeth 51, 52, 53, 55, and 16 were lost at post- or peri-mortem.
The development of the unerupted permanent teeth suggests an age of
4.5 - 5 years.
*F.D.I. notation
|
X-Rays are Shown in "Positive" and "Negative" Views
Go Back
Report from Trace Genetics
|

Report on the DNA
analysis from skeletal remains from two skulls August
12, 2003
Two sets of remains were received by Trace Genetics and
were processed for genetic analyses. The remains
consisted of two skulls presented by Mr. Lloyd Pye for
DNA analysis.
SAMPLING
Prior to attempts to extract DNA from the remains, the
remains were inventoried and taped using a video camera.
Video records of the sampling procedure and the initial
extraction on all samples were taken and archived by
Trace Genetics.
Samples were cut from the left parietal of an abnormally
shaped skull, identified as the Starchild skull on
February 10, 2003. Equipment used to sample was
sterilized using a bleach solution prior to use.
Sampling was performed in a room not used for any
genetic analyses. Fragments weighing a total of 0.8g
were cut from the parietal using a rotary cutter with a
previously unused blade. The fragments were placed in a
sterile conical tube labeled SCS-1 and stored for
analysis. A second 0.7g fragment adjacent to the sample
retained by Trace Genetics was placed in a sterile
conical tube labeled SCS-2 and returned to Mr. Pye.
Two teeth were removed from maxilla of a skull presented
in association with the Starchild skull on February 10,
2003. The right first molar tooth and root weighing 1.7g
was removed and labeled “SA-1.” The tooth and root were
placed in a sterile conical tube and retained for
genetic analysis by Trace Genetics. A portion of the
root was fractured in the process and remained in the
maxilla. The right premolar and root (sample labeled
“SA-2”; total weight 1.0g) were also removed from the
maxilla, placed in a sterile conical tube. The SA-2
sample was returned to Mr. Pye.
EXTRACTION AND ANALYSIS OF DNA SCS-1:
Extraction 1:
A first extraction was performed on a 0.24g
fragment of the parietal bone from sample SCS-1. The
extraction was performed in a dedicated ancient DNA
laboratory beginning on 7 March 2003 and was
performed in parallel to an extraction of SA-1 and a
reagent blank (negative control). Both surfaces of
bone were sanded with a rotary sander to remove any
surface contaminants and lacquer preserves present
on the outer surfaces of bone. Subsequent to
sanding, the bone was exposed to ultraviolet (UV)
irradiation (254nm) for 300 seconds per side. The
bone surface was then cleaned with bleach (2% sodium
hypochlorite), rinsed with sterile EDTA and placed
in a fresh 15ml conical tube and immersed in
approximately 2ml of 0.5M EDTA. The tube was sealed
with parafilm and placed on a rocker.
After 10 days, the tube was opened and 150µl of 0.1M
PTB [1] and 20µl of 100mg/ml proteinase-K was added
to the sample and EDTA. The sample was incubated
with agitation overnight at 64oC. DNA was extracted
from the digested sample using a 3-step
phenol/chloroform extraction method. Two extractions
with phenol:chloroform:isoamyl
(25:24:1) of equal volume to the digested product
were followed by an extraction with an equal volume
of chloroform:isoamyl (24:1). The extracted DNA
solution was concentrated by ammonium-acetate
precipitation using two volumes of cold 100%
filtered ethanol and 1/2 volume of 5M ammonium
acetate. This solution was then stored at -20�C for
approximately 4 hours to facilitate precipitation,
then centrifuged at high speeds (10,000-12,000rpm)
for 15 minutes to pellet the precipitated DNA. The
supernatant was discarded and the remaining DNA
pellet dried and resuspended in ~300µl sterile
ddH2O. To further purify the DNA and remove
additional PCR inhibitors co-extracted with the DNA,
the DNA solution was purified using the Promega�
Wizard PCR Preps DNA Purification Kit as directed by
the manufacturer. DNA was eluted from Promega�
columns with 100µl sterile ddH2O, the elutant
labeled SCSex1 and stored at –20�C.
Attempts to amplify segments of mtDNA from extract
SCSex1 were performed as described below in METHODS.
Single amplifications for fragments containing the
diagnostic mutations for Native American haplogroups
A, B, C and D[2] did not reveal a known Native
American haplogroup, however, the extraction did not
amplify consistently. A single amplification of a
fragment of the mtDNA first hypervariable segment (HVSI)
between np 16210 and np 16328 was sequenced using a
cycle sequencing procedure with ABI Big-Dye 3.1
chemistry and analyzed on an ABI automated genetic
analyzer. The sequence obtained revealed a
transition relative to the Cambridge reference
sequence at np16273. This sequence did not match
either any personnel with access to the ancient DNA
facilities or a sequence obtained from Mr. Pye.
Subsequent amplifications of this fragment were not
successful and the sequence could not be confirmed.
Attempts to amplify fragments of the amelogenin gene
located on the X and Y chromosome[3] were uniformly
not successful.
Extraction 2:
A second extraction was performed beginning
April 21, 2003 on 0.21g of the parietal sample from
SCS-1. The extraction was performed as above with
the following modifications:
-
The sample was run
in parallel with a reagent blank (negative
control) but was not processed with any other
samples.
-
The bone was exposed
to 900 seconds of UV irradiation per side.
-
The bone was
completely immersed in 2% sodium hypochlorite
for 5 minutes.
-
The sample was left
in EDTA with agitation for 22 days prior to
digestion with proteinase-K.
-
At digestion, ~50µl
of Tween-20 was added with 100µl of PTB.
-
The silica
extraction columns (Promega®) were eluted with
80µl of ddH2O and sample labeled “SCSe2.”
Attempts to amplify
mtDNA for fragments containing the diagnostic
mutations for Native American haplogroups A, B, C
and D were performed on extract SCSe2. Multiple
amplifications indicated that the sample possessed
an AluI restriction site at np 13262 indicative of
Native American haplogroup C [2]. Sequence obtained
for a fragment of the first hypervariable segment of
the mtDNA control region from np16210 to np 16367
revealed transitions at np16223, np 16298, np 16325
and np 16327. These mutations are characteristic of
haplogroup C in the Americas [4].
Multiple attempts to amplify a segment of the
amelogenin gene were unsuccessful using various
amounts of SCSe2 extract as template. 30µl of the
original extract was concentrated to a final volume
of ~10µl using a microcon YM-30 concentrator.
Attempts to amplify this concentrated template were
not successful.
Extraction 3:
A third extraction was performed beginning on
June 4, 2003 as described above for extraction 2
with the following modifications:
-
The extraction was
performed on the entire remaining 0.40g of bone.
-
The sample was
immersed in ~3.5ml of EDTA.
-
To ensure adequate
demineralization of the sample, the sample was
left immersed in EDTA with agitation for 30
days.
-
The final elution
from the silica spin columns (Promega®) was
performed twice, each time with 35µl of ddH2O
preheated to 65oC.
Attempts to amplify
fragments of mtDNA were performed to test for the
presence of diagnostic mutations fo r Native
American haplogroups A and C. The sample did not
appear to possess the diagnostic HaeIII mutation and
np663 indicative of haplogroup A. Multiple
amplifications did reveal the presence of the AluI
site gain at np13262 indicative of haplogroup C.
A single amplification of a fragment of the
amelogenin gene located on the X and Y chromosomes
[3] produced a single amplification product 106bp in
length. Multiple subsequent amplifications did not
reproduce this event, as all subsequent attempts did
not produce a PCR product.
SA-1:
Extraction 1:
A first extraction was performed on 0.53g
fragment of the molar tooth from sample SA-1
beginning on 7 March 2003 and was performed in
parallel to an extraction of SCS-1 (above) and a
reagent blank (negative control). The extraction was
performed in the manner describe above for
extraction 1 of SCS-1 save that the outer surface of
the tooth, which had previous to sampling been
firmly rooted in the maxilla, was not sanded and the
final elution of the silica spin column (Promega®)
was eluted to 100µl and labeled SAex1.
Multiple attempts to amplify segments of mtDNA
containing amplifications for fragments of mtDNA
containing the diagnostic mutations for Native
American haplogroup A revealed a HaeIII restriction
site at np663 consistent with known Native American
haplogroup A [2]. Amplifications for fragments
containing the diagnostic sites for haplogroups B, C
and D did not show presence of mutations indicative
of these haplogroups. A single amplification of a
fragment of the mtDNA first hypervariable segment (HVSI)
between np 16210 and np 16327 revealed transitions
relative to the Cambridge reference sequence at
np16223, np16290 and np16319. These mutations are
consistent with Native American haplogroup A.
Multiple amplifications of a fragment of the
amelogenin gene on the X and Y chromosomes
consistently produced a single band 106bp in length
when visualized on an electrophoretic gel consistent
with DNA from a female [3].
Extraction 2:
A second extraction was performed beginning Ap
ril 21, 2003 on 0.42g of the tooth sample from SA-1.
The extraction was performed similar to extraction 1
on SA-1 (above) with the following modifications:
Multiple amplifications
of a mtDNA fragment indicated the presence of a
HaeIII restriction site at np663 indicative of
Native American haplogroup A. Amplifications of the
extraction did not possess the AluI site gain at
np16262. Multiple amplifications of a fragment of
the amelogenin gene produced a single band when
visualized on an electrophoretic gel consistent with
DNA from a female.
DISCUSSION:
MtDNA from virtually all modern, full-blooded Native
Americans belongs to one of five mitochondrial lineages
or matrilines (designated haplogroups A, B, C, D, and X)
marked by the presence or absence of characteristic
restriction sites or by the presence of a nine base pair
(9-bp) deletion [2, 5]. Analyses of ancient DNA from
Native Americans likewise indicates that these
haplogroups constitute virtually all prehistoric Native
American individuals as well [see: 6].
American haplogroup C, as revealed through two
independent extractions performed on fragments of
parietal bone. While a single first extraction did not
appear to type similarly, this inconsistent result is
likely a product of a low level of contamination. This
single extraction neither amplified consistently nor was
the single sequence of HVSI reproducible. Contamination
could have occurred either prior to sampling, introduced
in the extraction process, or during PCR amplifications.
It is unlikely that contamination could account for the
haplogroup C mtDNA as this type is not possessed by any
researcher with access to the ancient DNA facilities and
the reagent blanks did not indicate systematic
contamination in the extractions.
The sample taken from the associated skull (SA-1) has
mtDNA consistent with Native American haplogroup A as
determined through both extractions. The sample also
appeared be from a female individual as evidenced by
repeated amelogenin typing. It is unlikely that
contamination could account for the haplogroup A mtDNA
as this type is not possessed by any researcher with
access to the ancient DNA facilities and the reagent
blanks did not indicate systematic contamination in the
extractions.
As mtDNA exists in high copy number (upwards of three
orders of magnitude relative to any single copy nuclear
DNA locus), it can be recovered from prehistoric
biological material in sufficient quantities for
amplification and analysis using the polymerase chain
reaction (PCR) [see: 7, 8]. MtDNA is present in haploid
condition with inheritance being passed down exclusively
through maternal lines [9]. Thus, that the samples
analyzed from SCS-1 and SA-1 possessed markedly
different mtDNA types excludes a mother-offspring
relationship between the two individuals. As it was
possible to type and confirm both to known pre-Columbian
mtDNA types found in the Americas, both individuals most
appear to have possessed Native American mothers.
While it is possible to obtain nuclear DNA as well from
ancient samples, the reduced copy-number at any
particular nuclear locus relative to mtDNA makes it less
likely that a particular extract will contain sufficient
DNA for the analysis of a nuclear genetic locus using
presently available PCR methods. The ability to amplify
nuclear DNA from the SA-1 extractions but not from the
SCS-1 extractions could be a product of any of a number
of factors. In ancient DNA analysis, success rates from
teeth are generally higher than from bone [10, 11].
Further, there is some indication that X-Ray exposure
damages and degrades DNA, which may have decreased the
quantity and quality of DNA available in the bone prior
to extraction.
The lone amplification using
the amelogenin primers on extract SCSe3 could not be
confirmed through additional amplifications and likely
indicates a sporadic contamination of a single PCR
reaction caused either by a female individual in the
laboratory or could have been introduced to laboratory
disposables (e.g. pipette tips, PCR reaction tubes).
Such contamination has been noted elsewhere [12] and
consequently, any conclusions drawn from the single
un-reproduced PCR reaction should not be taken as any
reliable indication as to the DNA present in the sample.
The presence of reliably typed mtDNA from SCS samples
does indicate that mtDNA is present in the bone. The
inability to analyze nuclear DNA indicates that such DNA
is either not present or present in sufficiently low
copy number to prevent PCR analysis using methods
available at the present time.
All reagents used to extract and amplify were first
tested to detect any DNA contamination and ancient DNA
facilities were cleaned using bleach to remove possible
sources of contamination. Further additional
contamination controls and precautions are described
below.
PCR amplifications of mtDNA were conducted in 25 µl
volumes using 4µl dNTPs (10mM), 2.5µl 10X PCR buffer (Gibco),
1.3µl BSA (20mg/ml), 0.75µl MgCl2, 0.2µl Platinum Taq
DNA polymerase (Gibco) 2 to 6 µl of DNA template and
sterile ddH20 sufficient to bring reaction volume to
25µl. After an initial 4-minute denaturation step at 94o
C to activate the hot start Taq, 40 PCR cycles were
performed consisting of a 94oC denaturing step, a
50-55oC annealing step (temperature depending on primers
utilized), and a 72oC extension step of 30 seconds each.
A final 3-minute extension at 72oC was added after the
last cycle. A portion of the amplification product
(~5µl) was run on a 6% polyacrylamide gel together with
a size standard ladder, stained with ethidium bromide
and photographed under UV light using a digital imager
(ISO 2000 imaging system, Alpha Innotech, San Leandro,
CA).
To assess the presence or
absence of diagnostic restriction sites, the remaining
20µl were incubated with 10 units of the appropriate
restriction enzyme overnight at 37o C, and then
subjected to electrophoresis in the manner previously
described. Primers used for amplification of these
segments and restriction enzymes used are shown in table
1.
Amelogenin amplifications [3] were attempted using 1, 3,
and 8 µl of DNA template in 25 µl reaction volumes,
adjusting ddH2O amounts to maintain concentrations of
other reagents.
| Site | First Primer Coordinate | Second Primer Coordinate | Polymorphism | Reference: |
|---|
| Haplogroup A | 591-611 | 765-743 | HaeIII np663 “+” = A
| [13] |
|---|
| Haplogroup B | 8195-8215 | 8316-8297 | 9 base pair deletion Deletion = B | [14] |
|---|
| Haplogroup C | 13236-13257 | 13310-13290 | AluI np13262 “+” = C
| [15] |
|---|
| Haplogroup D | 5099-5120 | 5211-5190 | AluI np 5176 “-“ = D
| [15, 16] |
|---|
| HVSI | 16192-16209 | 16385-16368 | Sequence | [15, 17] |
|---|
| HVSI | 16192-16209 | 16348-16328 | Sequence | [15, 17] |
|---|
CONTAMINATION CONTROLS:
Ancient DNA is typically highly degraded and
survives in much lower copy numbers than modern DNA.
Consequently, ancient DNA is highly vulnerable to
contamination from modern sources and specific
precautions against contamination, as summarized by
Kelman and Kelman [18], were utilized in this study to
both minimize contamination and, importantly, to
identify contamination when present so that it does not
lead to false inferences. These measures include: 1. Use
of dedicated laboratory space, supplies, reagents and
equipment for preparation of ancient DNA samples inside
UV irradiated glove boxes; 2. Use of sterile, disposable
labware irradiation and bleaching of all materials used
to help eliminate any surface contamination; 5. Running
negative controls at all stages of the extraction and
amplification process to identify the presence of
contaminants; 6. Confirmation of results by multiple
amplifications of multiple extractions. All positive
results were confirmed though multiple amplifications of
each extraction and multiple extractions performed at
different times.
References Cited
-
Poinar, H.N., et al.,
Molecular coproscopy: Dung and diet of the extinct
ground sloth Nothrotheriops shastensis. Science
(Washington D C), 1998. 281(5375): p. 402-406.
-
Schurr, T.G., et al.,
Amerindian mitochondrial DNAs have rare Asian
mutations at high frequencies, suggesting they
derived from four primary maternal lineages.
American Journal of Human Genetics, 1990. 46(3): p.
613-23.
-
Mannucci, A., et al.,
Forensic Application of a Rapid and Quantitative DNA
Sex Test by Amplification of the X-Y Homologous Gene
Amelogenin. International Journal of Legal Medicine,
1994. 106(4): p. 190-193.
-
Torroni, A., et al.,
Asian affinities and continental radiation of the
four founding Native American mtDNAs. American
Journal of Human Genetics, 1993. 53(3): p. 563-590.
-
Forster, P., et al.,
Origin and evolution of native American mDNA
variation: A reappraisal. American Journal of Human
Genetics, 1996. 59(4): p. 935-945.
-
Eshleman, J.A., R.S.
Malhi, and D.G. Smith, Mitochondrial DNA Studies of
Native Americans: Conceptions and Misconceptions of
the Population Prehistory of the Americas.
Evolutionary Anthropology, 2003. 12(1): p. 7-18.
-
Kaestle, F.A. and D.G.
Smith, Ancient mitochondrial DNA evidence for
prehistoric population movement: The Numic
expansion. American Journal of Physical
Anthropology, 2001. 115(1): p. 1-12.
-
Stone, A.C. and M.
Stoneking, mtDNA analysis of a prehistoric Oneota
population: Implications for the peopling of the New
World. American Journal of Human Genetics, 1998. 62:
p. 1153-1170.
-
Giles, R.E., et al.,
Maternal Inheritance of human mitochondrial DNA.
Proceedings of the National Academy of Sciences of
the United States of America, 1980. 77(11): p.
6715-6719.
-
Malhi, R.S.,
Investigating prehistoric population movements in
North America with ancient and modern mtDNA, in
Anthropology. 2001, University of California: Davis.
-
Hummel, S., Ancient DNA
Typing: methods, strategies and applications. 2003,
Berlin: Springer-Verlag.
-
Schmidt, T., S. Hummel,
and B. Herrmann, Evidence of contamination in PCR
laboratory disposables. Naturwissenschaften, 1995.
82(9): p. 423-431.
-
Stone, A.C. and M.
Stoneking, Ancient DNA from a pre-Columbian
Amerindian population. American Journal of Physical
Anthropology, 1993. 92(4): p. 463-471.
-
Wrischnik, L.A., et al.,
Length mutations in human mitochondrial DNA: direct
sequencing of enzymatically amplified DNA. Nucleic
Acids Research, 1987. 15: p. 529542.
-
Handt, O., et al., The
retrieval of ancient human DNA sequences. American
Journal of Human Genetics, 1996. 59(2): p. 368-376.
-
Parr, R.L., S.W.
Carlyle, and D.H. O'Rourke, Ancient DNA analysis of
Fremont Amerindians of the Great Salt Lake Wetlands.
American Journal of Physical Anthropology, 1996.
99(4): p. 507-518.
-
Eshleman, J.A.,
Mitochondrial DNA and prehistoric population
movements in Western North America, in Anthropology.
2002, University of California, Davis: Davis, CA.
-
Kelman, L.M. and Z.
Kelman, The use of ancient DNA in paleontological
studies. Journal of Vertebrate Paleontology, 1999.
19(1): p. 8-20.
|
Go Back
|